
CERMAV | CNRS | PIG | SIB
Why UniLectin ?
The next frontier in understanding protein function is deciphering the “PTM code”. Glycosylation as the most variable posttranslational modification complicates the picture with its own challenging glycocode borne by eukaryotic surface glycoconjugates or microbial membrane polysaccharides. The ability of lectins to non-covalently bind glycans makes these proteins the key readers of information encoded by glycans. The roles of glycans and glycoconjugates are manifold and revealed in various medical, biochemical and biotechnological applications. Glycoscience is now reaching out to other omics and systems biology following recent technological progress in carbohydrate structure resolution and ab initio synthesis as well as functional screening methods. Glycoinformatics also expands in support of this growing knowledge as summarised in Glyco@Expasy, our overarching project.
Lectins are diverse within and across all species. This particular class of proteins is considered as a major player in many biological processes. Glycan-binding is based on multivalence, which gives lectins a unique capacity to interact with surface glycans and significantly contribute to cell-cell recognition and interactions. Lectins have been studied for many years using a broad range of technologies, nonetheless, they are poorly annotated in genomes and no reference database was available before the launching of the UniLectin platform in 2019. Structuring lectin knowledge was hitherto challenged by the lack of a reliable classification resisting to the continuous production of new sequences and the slow elucidation of corresponding 3D structures. The initial content of UniLectin consisted of manually curated lectin 3D structures together with their interacting glycans. In 2020, accumulated data set the basis of a robust hierarchical classification built on 35 protein folds and expanding into 109 classes. The latter were used to define 109 HMM profiles and extensively screen NCBI-nr and UniProt. This resulted in creating LectomeXplore that includes ~106 lectin predictions across all kingdoms. Species distribution of lectins appears wider than expected. LectomeXplore is the first resource supporting lectin annotation and bolsters the success of UniLectin recognised as a key step toward breaking the glycocode.
- For a practical approach to UniLectin, see this bioinformatics protocol: Imberty A, Bonnardel F, Lisacek F. UniLectin, A One-Stop-Shop to Explore and Study Carbohydrate-Binding Proteins. Curr Protoc. 2021 Nov;1(11):e305. doi: 10.1002/cpz1.305. PMID: 34826352
UniLectin3D
UniLectin3D includes manually curated structural information on lectins along with their interactions with carbohydrate ligands. A classification is proposed based on taxonomic origin and structure fold. This family-based classification cross-links to other glyco-related databases of the Glyco@Expasy collection. The content of UniLectin3D is centered on three-dimensional data, using PDB information, with an appropriate curation of the glycan topology. The 3D visualization of contacts between the lectin and the ligand is performed via the Protein-Ligand Interaction Profiler (PLIP) applet of the Swiss-Model repository.
Further details can be found in:
- Bonnardel F, Perez S, Lisacek F, Imberty A. Structural Database for Lectins and the UniLectin Web Platform, Methods Mol Biol. 2020;2132:1-14. 10.1007/978-1-0716-0430-4_1. PMID: 32306309
- Bonnardel F, Mariethoz J, Salentin S, Robin X, Schroeder M, Perez S, Lisacek F, Imberty A. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands, Nucleic Acids Res. 2019 Jan 8;47(D1):D1236-D1244. 10.1093/nar/gky832. PMID: 30239928
LectomeXplore
LectomeXplore describes candidate lectins identified in available proteomes from all kingdoms based on the classification of UniLectin3D composed of 109 lectin classes. LectomeXplore contains the results of screening yearly the UniProt (Swiss-Prot + TrEMBL) and NCBI-nr (non-redundant) protein databases to identify lectin candidates with 109 tailormade Hidden Markov Model (HMM) profiles, with at least 50% of sequence similarity to the reference motif.
Predicted lectins are unique to the UniLectin platform and open the way to running lectin profiles of LectomeXplore to search genomes and reveal specific lectomes. Our first attempt involved multiple bacteria of the vaginal microbiome. Pathogenic species causing adverse pregnancy outcomes were shown to be enriched in a larger diversity of lectins than commensal species. This supports a broader capacity of pathogenic microbes to attach to the host glycome.
Further details can be found in:
- Bonnardel F, Mariethoz J, Pérez S, Imberty A, Lisacek F. LectomeXplore, an update of UniLectin for the discovery of carbohydrate-binding proteins based on a new lectin classification, Nucleic Acids Res. 2021 Jan 8;49(D1):D1548-D1554. 10.1093/nar/gkaa1019. PMID: 33174598
- Bonnardel F, Haslam SM, Dell A, Feizi T, Liu Y, Tajadura-Ortega V, Akune Y, Sykes L, Bennett PR, MacIntyre DA, Lisacek F, Imberty A. Proteome-wide prediction of bacterial carbohydrate-binding proteins as a tool for understanding commensal and pathogen colonisation of the vaginal microbiome, NPJ Biofilms Microbiomes. 2021 Jun 15;7(1):49. 10.1038/s41522-021-00220-9. PMID: 34131152
BiotechLec
For decades, lectins have been used as probes in glycobiology and this usage has gradually spread to other domains of Life Science. Nowadays, researchers investigate glycan recognition with lectins in diverse biotechnology and clinical applications, addressing key questions regarding binding specificity. The latter is documented in scattered and heterogeneous sources, and this situation calls for a centralised and easy-access reference. BiotechLec was designed to address this need.
Further details can be found in:
- Schnider B., Lorenzo-Escudero F., Imberty A., Lisacek F. BiotechLec: an interactive guide of commercial lectins for glycobiology and biomedical research applications, Glycobiology, Volume 33, Issue 9, September 2023, Pages 684–686. 10.1093/glycob/cwad034. PMID: 37083961
HumanLectome
Human lectins have various functions that can be related to their localisation. For instance, intracellular lectins are mostly involved in quality control of glycoprotein biosynthesis and intracellular trafficking. The quality control is also at play on the surface of cells where asialoglycoprotein receptors (ASGPR) on mammalian hepatocytes are involved in the turnover of serum glycoproteins. Through binding to endogenous glycans, cell surface lectins participate in cell–cell and cell–matrix interactions. Furthermore, human lectins are key players in innate immunity by recognising non-self glycans on viruses, bacteria, parasites and fungi. Soluble lectins in serum activate a variety of defence mechanisms, from phagocytose to activation of the complement cascade. Lectins on immune cells possess intracellular signalling domains and are involved in activation and repression of immunity responses. This variety of functions and localisations is mirrored by the structural variety of Carbohydrate Recognition Domains (CRDs), but also by a large range of architectures. Many lectins are composed of a single CRD, that can associate as dimers or oligomers, while others are part of complex multi-domain proteins that may be anchored to the plasma membrane to exert further signalling function(s). HumanLectome covers knowledge of the complete set of human lectins as reported in the literature or suspected/predicted with limited evidence.
Further details can be found in:
- Schnider B., M'Rad Y., el Ahmadie J., de Brevern AG., Imberty A., Lisacek F. HumanLectome, an update of UniLectin for the annotation and prediction of human lectins, Nucleic Acids Reasearch. 10.1093/nar/gkad905. PMID: 37889052
PropLec
PropLec includes known and predicted β-propeller lectins along with their features and conserved regions. A β-propeller is defined as a particular type of all β-protein architecture characterized by four to eight highly symmetrical blade-shaped β-sheets arranged toroidally around a central axis. Together the β-sheets form a funnel-like active site. The blade consists of a small domain of less than 50 amino acids. The repeated blades hamper the identification of similar lectins when using common software based on pairwise sequence alignment. However, the multiple alignment of blades manually adjusted with knowledge of 3D structures produces a unique conserved domain as illustrated below. This blade domain can then be used to compare all known β-propeller lectins and this systematic comparison led to the definition of five distinct families. The specific signature of each family has been used to predict possible β-propeller lectins from the UniProt (Swiss-Prot + TrEMBL) and NCBI-nr (non-redundant) protein databases. Results are directly accessible in PropLec and also included in the LectomeXplore module.
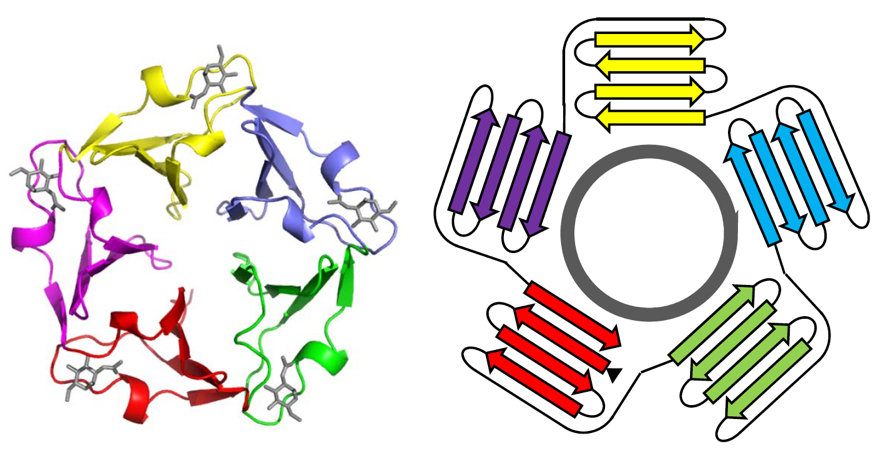
Five-bladed tachylectin-2 (1TL2) as an example of β-propeller and its schematic representation
Further details can be found in:
- Bonnardel F, Kumar A, Wimmerova M, Lahmann M, Perez S, Varrot A, Lisacek F, Imberty A. Architecture and Evolution of Blade Assembly in β-propeller Lectins, Structure. 2019 May 7;27(5):764-775.e3. 10.1016/j.str.2019.02.002. PMID: 30853410
- Notova S, Bonnardel F, Lisacek F, Varrot A, Imberty A. Structure and engineering of tandem repeat lectins, Curr Opin Struct Biol. 2020 Jun;62:39-47. 10.1016/j.sbi.2019.11.006. PMID: 31841833
TrefLec
β-trefoil lectins are popular in protein engineering for the design of new scaffolds with high symmetry that can be associated with other domains. For example, β-trefoils of the Ricin-like lectin class are associated with glycosyl hydrolases or lipases, while the β-trefoil Cys-rich domain is part of the macrophage mannose receptor that also contains fibronectin and multiple C-type protein domains. Following the same principle mentioned for β-propeller lectins, tandem repeats of β-trefoils were aligned with the knowledge of 3D structures to create as precise as possible HMM profiles. These models were used to screen the UniProt (Swiss-Prot + TrEMBL) and NCBI-nr (non-redundant) protein databases. Results are directly accessible in TrefLec and also included in the LectomeXplore module. Further investigation of predicted β-trefoil lectins in association with an aerolysin domain led to identify a pore forming toxin in a colony-forming micro-eukaryotic marine organism.
Further details can be found in:
- Lebreton A, Bonnardel F, Dai YC, Imberty A, Martin FM, Lisacek F. A Comprehensive Phylogenetic and Bioinformatics Survey of Lectins in the Fungal Kingdom, J Fungi (Basel). 2021 Jun 7;7(6):453. 10.3390/jof7060453. PMID: 34200153
MycoLec
A large number of lectins have been identified in filamentous fungi and yeasts. They are generally considered as defense-related proteins. This role is well documented in mushroom-forming fungi, where lectins are known to protect these reproductive structures from hyphal grazers, such as nematodes, slugs, snails and insects. That is why fungal lectins are of potential interest in agriculture as entomotoxic and nematotoxic compounds. Furthermore, some fungal lectins specifically bind oligosaccharide epitopes on cancer cells and are therefore promising tools for diagnosis of drug release. MycoLec contains the prediction of 33,518 fungal lectins belonging to 63 distinct lectin classes from 1419 genomes of the MycoCosm database. Significant differences in the lectomes of translated genomes support the distinction across fungal taxonomic classes. Moreover, lectin occurrence can be correlated with ecological information available in selected fungal species.
Further details can be found in:
- Notova S, Bonnardel F, Rosato F, Siukstaite L, Schwaiger J, Lim JH, Bovin N, Varrot A, Ogawa Y, Römer W, Lisacek F, Imberty A. The choanoflagellate pore-forming lectin SaroL-1 punches holes in cancer cells by targeting the tumor-related glycosphingolipid Gb3, Commun Biol. 2022 Sep 12;5(1):954. 10.1038/s42003-022-03869-w. PMID: 36097056
By using this site you agree to our privacy policy.
Please confirm you agree with the privacy policy before using the site.
 How to cite
How to cite